Transposable elements (TEs), also known as “jumping genes,” are DNA sequences that can change their position within a genome. These elements are present in almost all organisms, including plants, animals, and even bacteria. The discovery of transposable elements revolutionized our understanding of genetics, showing that the genome is not a static entity but rather a dynamic system that can change over time.
Table of Contents
What are Transposable Elements?
Transposable elements are segments of DNA that have the ability to move (transpose) from one location to another within the genome. This movement can occur either by cutting and pasting the element to a new location or by copying the element and inserting the copy elsewhere. The process of movement is known as transposition.
Transposable elements were first discovered by Barbara McClintock in the 1940s during her work on maize (corn). Her discovery challenged the traditional view that genes were fixed in place within chromosomes and earned her the Nobel Prize in Physiology or Medicine in 1983.

Characteristics of Transposable Elements
There are several key characteristics of transposable elements:
Mobility
The defining feature of TEs is their ability to move from one genomic location to another. This movement can be random, meaning that the TE can insert itself into various locations within the genome.
Types of Transposable Elements
Transposable elements are broadly classified into two major types:
Class I elements (Retrotransposons)
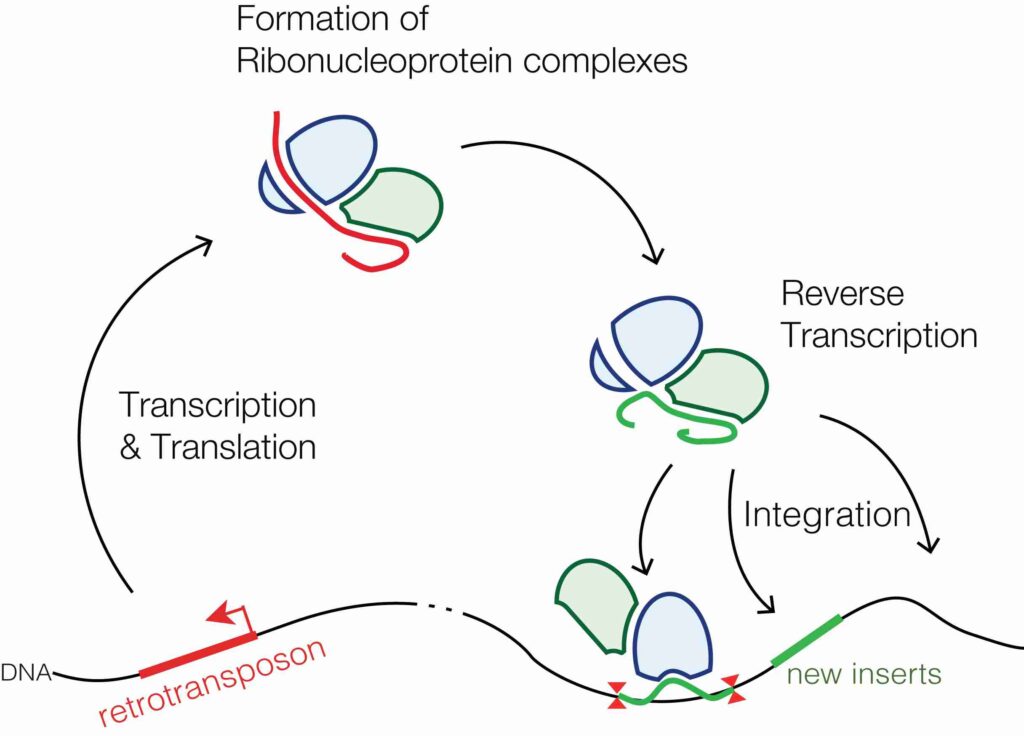
These elements move via a “copy and paste” mechanism, meaning they are first transcribed into RNA, and then the RNA is reverse-transcribed back into DNA before being inserted into a new location. Retrotransposons often leave behind a copy of themselves at the original location.
Class II elements (DNA transposons)
These elements move via a “cut and paste” mechanism. A DNA transposon is excised from its original location and inserted into a new site without creating a copy.
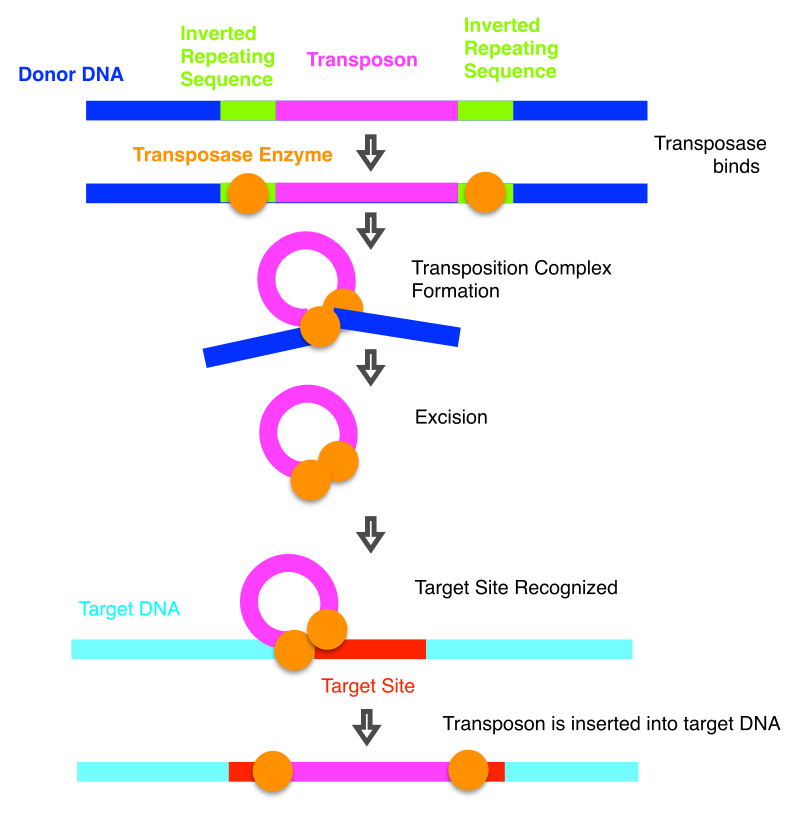
Autonomous and Non-Autonomous Elements:
Autonomous transposable elements
It carry all the machinery necessary for their own movement, including genes that encode enzymes (like transposase or reverse transcriptase) required for transposition.
Non-autonomous transposable element
It lack the genes needed for transposition and depend on enzymes produced by autonomous elements to move.
Presence in the Genome
Transposable elements can make up a significant portion of an organism’s genome. For instance, in humans, nearly 45% of the genome consists of transposable elements, while in some plants like maize, TEs make up more than 80% of the genome.
Genomic Impact
TEs can cause mutations if they insert themselves into or near functional genes. They can disrupt gene expression, alter gene regulation, or even cause chromosomal rearrangements. However, they can also contribute to genome evolution by creating genetic diversity.
Mechanisms of Transposition
The process by which transposable elements move within the genome is called transposition. There are two main mechanisms of transposition:
Class I (Retrotransposons) – Copy and Paste Mechanism
Retrotransposons move by a process that involves first being transcribed from DNA into RNA and then reverse-transcribed back into DNA. This newly created DNA copy is inserted into a new genomic location.
Step 1: Transcription: A retrotransposon is transcribed into RNA by the cell’s machinery.
Step 2: Reverse Transcription: The RNA is reverse-transcribed back into DNA by an enzyme called reverse transcriptase, which is often encoded by the retrotransposon itself.
Step 3: Insertion: The new DNA copy is integrated into a different part of the genome by an enzyme called integrase.
Since the original retrotransposon remains in place and a new copy is inserted elsewhere, this process increases the number of copies of the element in the genome. Over time, retrotransposons can accumulate and make up a large portion of the genome.
Class II (DNA Transposons) – Cut and Paste Mechanism
DNA transposons move by directly excising themselves from one location and inserting into a new site. The key enzyme involved in this process is transposase, which recognizes specific DNA sequences at the ends of the transposon and catalyzes the movement.
Step 1: Excision: The transposase enzyme cuts the transposon out of its original location in the genome.
Step 2: Integration: The transposon is inserted into a new location within the genome. The transposase enzyme also catalyzes the integration of the transposon into the target DNA.
Unlike retrotransposons, DNA transposons typically do not increase in number because the transposon physically moves from one location to another without creating a copy.
Effects of Transposition on the Genome
Transposition can have several effects on the genome, both positive and negative. These effects contribute to the dynamic nature of genomes and play a role in evolution.
Mutagenesis: Transposable elements can disrupt genes or regulatory sequences when they insert themselves into or near important genomic regions. This can lead to mutations that may cause diseases or alter the function of genes. For example, in humans, some TEs are associated with genetic disorders like hemophilia and muscular dystrophy.
Gene Duplication and Rearrangement: TEs can promote genetic diversity by causing duplications, deletions, and rearrangements of genetic material. For instance, they can cause gene duplications by inserting near a gene and carrying it along when they move. These duplications can then evolve into new genes with different functions, contributing to the complexity of genomes.
Regulation of Gene Expression: Some transposable elements carry regulatory sequences that can influence the expression of nearby genes. In some cases, TEs can provide new promoters or enhancers that regulate when and where genes are expressed. This can lead to the evolution of new gene regulatory networks.
Genome Size: The accumulation of transposable elements over time can lead to an increase in genome size, a phenomenon known as genome expansion. For example, much of the large genome size in plants like maize is due to the presence of numerous transposable elements.
Defense Mechanisms: Organisms have evolved mechanisms to control and limit the activity of transposable elements, as unchecked transposition could cause genomic instability. One such defense mechanism is the RNA interference (RNAi) pathway, where small RNAs target TEs for degradation or suppression, preventing their movement.
Examples of Transposable Elements
LINEs (Long Interspersed Nuclear Elements): These are a type of retrotransposon found in the genomes of many organisms, including humans. LINEs are autonomous elements that can replicate and move by encoding reverse transcriptase and other necessary enzymes.
SINEs (Short Interspersed Nuclear Elements): These are non-autonomous retrotransposons that rely on the enzymes produced by LINEs to transpose. The Alu element is a well-known SINE in the human genome.
P Elements: In fruit flies (Drosophila), P elements are DNA transposons that can cause hybrid dysgenesis, a condition characterized by high rates of mutation and sterility in offspring.
Role of Transposable Elements in Evolution
Transposable elements have played a significant role in the evolution of genomes. Their ability to move, create mutations, and cause gene duplications has contributed to genetic diversity and adaptation. Some researchers believe that TEs may have been crucial in the evolution of complex organisms by driving the development of new genes and regulatory systems.
For instance, retrotransposons have been linked to the evolution of the mammalian placenta. Some TEs contain genes that are important for cell fusion, a process necessary for the development of the placenta.
Conclusion
Transposable elements are fascinating components of the genome that challenge our traditional understanding of genetic stability. Their ability to move, replicate, and influence genome structure has far-reaching implications for evolution, genetic diversity, and even disease. While they can sometimes cause harmful mutations, they also drive innovation within the genome by promoting genetic diversity and complexity. Understanding transposable elements provides insight into how genomes evolve and adapt over time.
Frequently Asked Questions (FAQ)
Define Mobility?
Mobility refers to the ability to move or be moved freely and easily from one place to another.
What do you mean by DNA transposons?
DNA transposons, also known as “jumping genes,” are segments of DNA that can move from one location to another within a genome. They do this through a “cut and paste” mechanism, where the DNA transposon is removed from its original spot and inserted into a new location.
Related Articles