What do you mean by DNA Microarray?
In molecular biology, DNA microarrays, sometimes referred to as gene chips, are potent instruments that enable scientists to test thousands of genes’ expression levels at once. In essence, they are tiny labs on a chip that can produce enormous volumes of information regarding the activity of genes in a cell or tissue. Understanding intricate biological processes, locating illness biomarkers, and creating fresh approaches to diagnosis and treatment are all made possible by this knowledge.
Table of Contents
The Principle of DNA Microarrays
Fundamental to DNA microarray technology is the process of hybridization, which is the binding of two complementary strands of DNA to one another.
Extracting Target RNA
RNA extraction from the target cells or tissues is the first step in the procedure. The sample’s current gene expression profile is represented by this RNA.
Transcription in reverse and Labeling
Reverse transcriptase is then used to transform the RNA into complementary DNA (cDNA). Fluorescent dyes, usually Cy3 (green) or Cy5 (red), are used to mark the cDNA in order to make certain sequences visible.
Blending
Following labeling, the cDNA is placed on a microarray slide that has thousands of known DNA sequences (probes) positioned at particular points. By binding to the array’s complementary sequences, the cDNA will produce a “hybridization pattern.”
Data Analysis and Scanning
A laser is used to scan the microarray slide in order to find the fluorescent signals that are produced by the attached cDNA. The abundance of the matching mRNA in the original sample is reflected in the signal intensity at each site. Next, this data is subjected to specialist software analysis in order to discover genes that exhibit differential expression in various samples or situations.
Types of DNA Microarrays
There are two main types of DNA Microarrays :
cDNA microarrays
These microarrays use cDNA probes, which are usually expressed sequence tags (ESTs) or cloned genes. When examining gene expression in a particular tissue or cell type, they are especially helpful.
Enzyme-Based Microarrays
Compared to cDNA microarrays, these arrays have improved specificity and sensitivity because they use shorter DNA sequences, or oligonucleotides, as probes. They are useful for identifying minute variations in gene expression and for investigating gene expression at a higher resolution.
Steps Involved in cDNA Microarrays
The process of performing a cDNA microarray experiment can be summarized in the following steps,
Preparing the Sample
RNA Isolation: Use specialized techniques, such as Trizol reagent, to extract RNA from the relevant samples. High quality and quantity of RNA are ensured by this procedure.
Quality Control: Determine the integrity of the RNA by employing techniques such as RNA integrity number (RIN) analysis or electrophoresis. This guarantees that the RNA is ready for additional processing.
Reverse Transcription: Use reverse transcriptase to transform the RNA into cDNA. For the molecule to be stable and easily detectable, this step is essential.
Labeling: During the reverse transcription process, add fluorescent dyes to the cDNA. The sort of analysis being done and the experimental design will determine which dye is used.
Hybridization using Microarrays
Array Preparation: The microarray slide should be prepared by adhering particular DNA probes to its surface. These probes stand in for transcripts or genes that are known.
Hybridization: Place the microarray slide and the labeled cDNA in an environment that promotes hybridization. As a result, the cDNA can attach to the array’s complementary sequences.
Cleaning: Clean the microarray slide to get rid of unbound cDNA and cut down on background interference. By doing this, the data quality and signal-to-noise ratio are both improved.
Data analysis and scanning
Scanning: To find the fluorescent signals from the attached cDNA, use a laser scanner to scan the microarray slide. The abundance of the matching mRNA in the original sample is represented by the signal intensity at each point.
Extraction of Data: Take the unprocessed data out of the scanner. Usually, the intensity values for every place on the array make up this data.
Normalization: Adjust the data for differences in labeling effectiveness and additional experimental variables. Ensuring reliable comparison of gene expression across samples is ensured by this step.
Analysis: To find genes and pathways that are differentially expressed, analyze the normalized data using statistical techniques and specialized tools. This stage provides biological information about the sample and how it reacts to the test setup.
Advantages and Applications of DNA Microarrays
DNA Microarrays offer several advantages:
High Throughput: They provide a thorough understanding of gene expression by enabling the simultaneous investigation of hundreds of genes.
Sensitivity: They are capable of picking up on minute variations in gene expression that other techniques might miss.
Versatility: They are useful for researching a variety of biological processes, including as drug response, illness, and development.
DNA Microarrays have numerous applications in research, diagnostics, and drug development, including:
Disease Diagnosis: It recognizing certain patterns of gene expression linked to various illnesses, which helps with early diagnosis and prognosis.

Drug discovery: It is the process of identifying targets, developing new pharmaceuticals, and assessing how medications affect gene expression.
Studies on Gene Regulation: Examining the processes that underlie gene regulation and determining the essential regulatory components.
Cancer research: Cancer research aims to comprehend the molecular causes of the disease, find biomarkers for diagnosis and therapy, and provide tailored treatments.
Environmental Toxicology: Determining how environmental contaminants affect gene expression and comprehending the harmful processes behind them.
The restricted uses of DNA microarrays
DNA microarrays are strong, yet they have several drawbacks.
Cost: Microarray experiments may be expensive, particularly when doing large-scale research.
Information Analysis Complexity: The enormous volume of data produced by microarrays necessitates the use of specialist software and knowledge for processing.
Cross-Hybridization: False positives may result from non-specific hybridization.
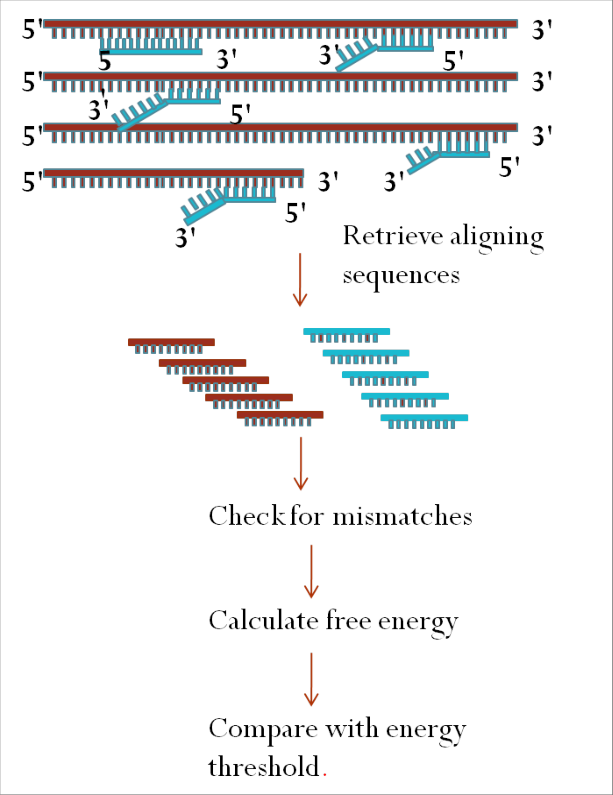
Limited Transcript Detection: Low-abundance transcripts may not be detected by microarrays due to their lack of sensitivity.
In summary
DNA microarrays have transformed our knowledge of gene expression and created new avenues for investigation and use in a variety of sectors. They are still a useful tool for researching the intricate interactions between genes and how they function in biological processes, despite several limitations. Microarrays are always being developed and enhanced as technology progresses, resulting in even higher sensitivity and resolution. DNA microarrays have the potential to contribute significantly to the advancement of life science and the creation of novel therapeutic and diagnostic approaches in the future.
Frequently Asked Questsions(FAQ)
What do you mean by therapeutic?
The word “therapeutic” is often used in relation to medicine and psychology, but it may also refer to anything that fosters wellbeing in a more general sense.
What are the types of DNA Microarrays?
The types of DNA Microarrays
1. cDNA microarrays
2. Enzyme – Based Microarrays
Related Articles