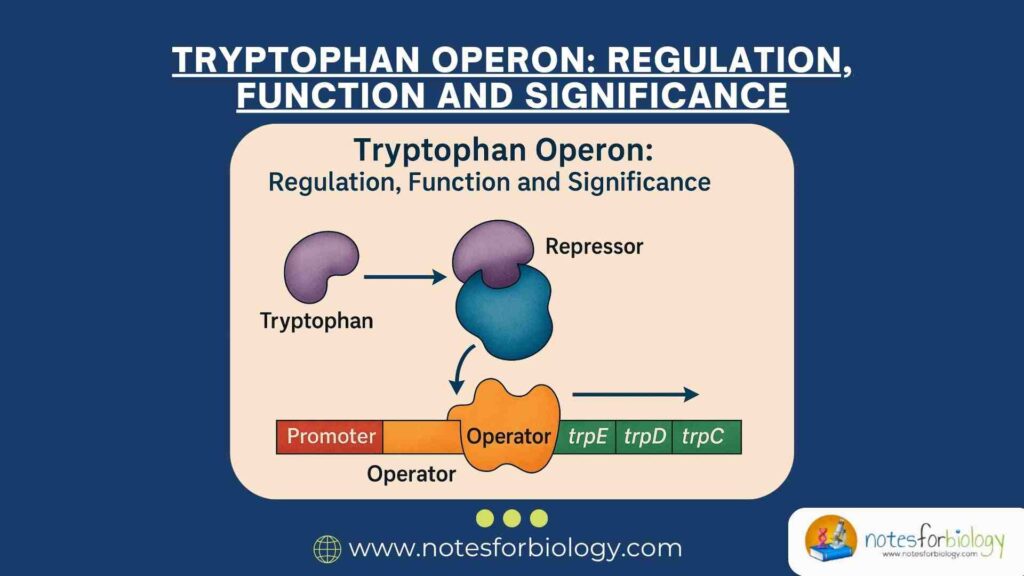
Gene regulation is a fundamental biological process that allows organisms to control the expression of their genetic information in response to environmental conditions. In prokaryotes, gene expression is often regulated through operons clusters of functionally related genes controlled by a single regulatory system. One of the best-characterized examples of this regulatory strategy is the Tryptophan (trp) operon in Escherichia coli (E. coli), a classic model for studying negative feedback inhibition and gene repression mechanisms.
The tryptophan operon is a cluster of structural genes in prokaryotic cells, particularly in E. coli, involved in the biosynthesis of the amino acid tryptophan. It operates under a negative feedback control mechanism, where the product of the operon, tryptophan, acts as a corepressor to inhibit further production when it is present in sufficient amounts. This operon serves as a classic example of a repressible operon system in prokaryotes.
Summary of Tryptophan Operon
- The tryptophan operon is a repressible operon in E. coli that controls enzyme production for tryptophan biosynthesis.
- Excess tryptophan binds the trp repressor, which attaches to the operator and blocks transcription.
- An attenuation mechanism at the leader sequence further fine-tunes transcription based on tryptophan levels.
Table of Contents
Location of the Tryptophan Operon
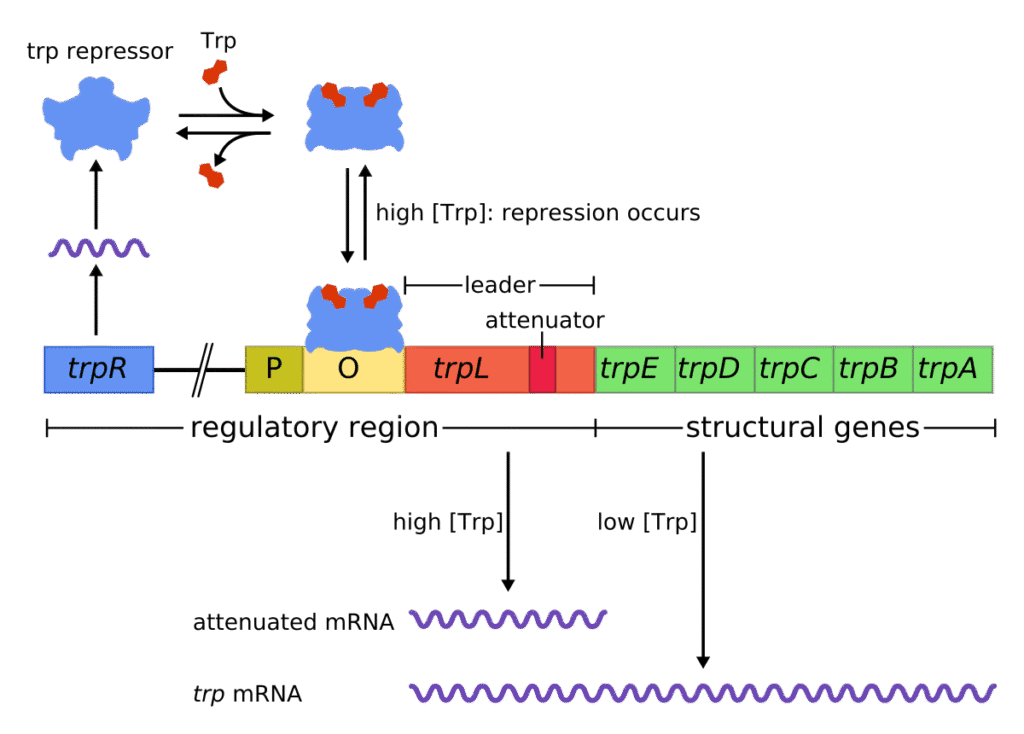
The tryptophan operon is located in the chromosomal DNA of prokaryotic organisms, most notably in E. coli. It is a continuous stretch of DNA that contains structural genes required for the biosynthesis of the amino acid tryptophan. In E. coli, it is positioned on the bacterial chromosome, and its exact chromosomal locus has been mapped through genetic studies, though the precise base pair numbering varies depending on the strain.
In general, operons like the trp operon are found in prokaryotic genomes because of their polycistronic nature, meaning a single mRNA molecule transcribed from the operon can translate into multiple proteins. This organization is uncommon in eukaryotic genomes, where genes are usually transcribed individually.
Components of the Tryptophan Operon
Structural Genes
The trp operon comprises five structural genes: trpE, trpD, trpC, trpB, and trpA. These genes encode the enzymes necessary for converting chorismic acid, a metabolic intermediate, into tryptophan through a multi-step biochemical pathway. Each gene contributes to producing a specific enzyme or enzyme subunit involved in the synthesis of tryptophan.
Promoter
Located upstream of the structural genes is the promoter region. It serves as the binding site for RNA polymerase, the enzyme responsible for initiating the transcription of the operon’s genes. The promoter ensures that the entire operon is transcribed as a single polycistronic mRNA when the operon is active.
Operator
Adjacent to the promoter is the operator sequence, a short stretch of DNA that interacts with the repressor protein. The operator functions as a regulatory element that can block the binding and progress of RNA polymerase when the repressor protein is bound to it, thereby preventing transcription of the structural genes.
Regulatory Gene (trpR)
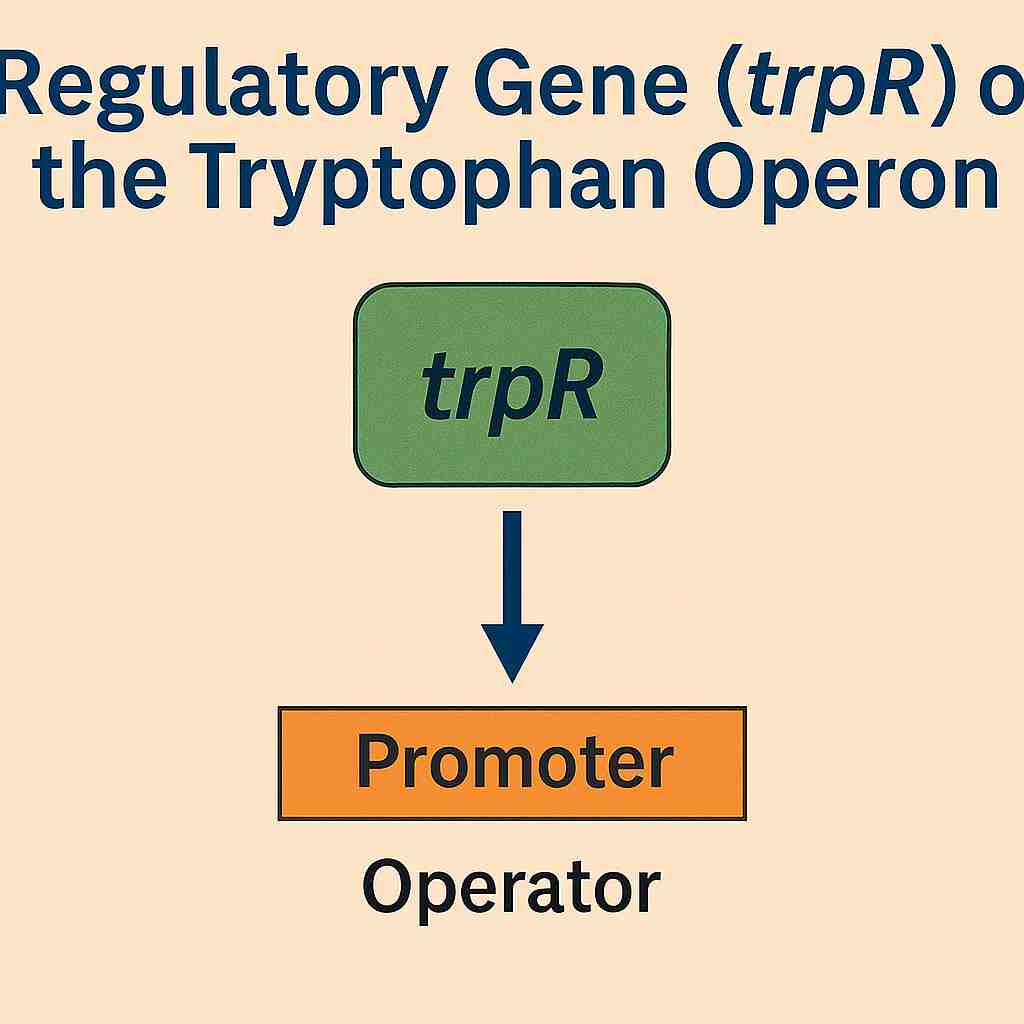
Separate from the operon itself, the trpR gene encodes the trp repressor protein. This gene is continuously expressed at a low level, producing inactive repressor proteins that can be activated by the binding of tryptophan molecules. Once activated, the repressor protein binds to the operator, repressing the transcription of the operon.
Leader Sequence
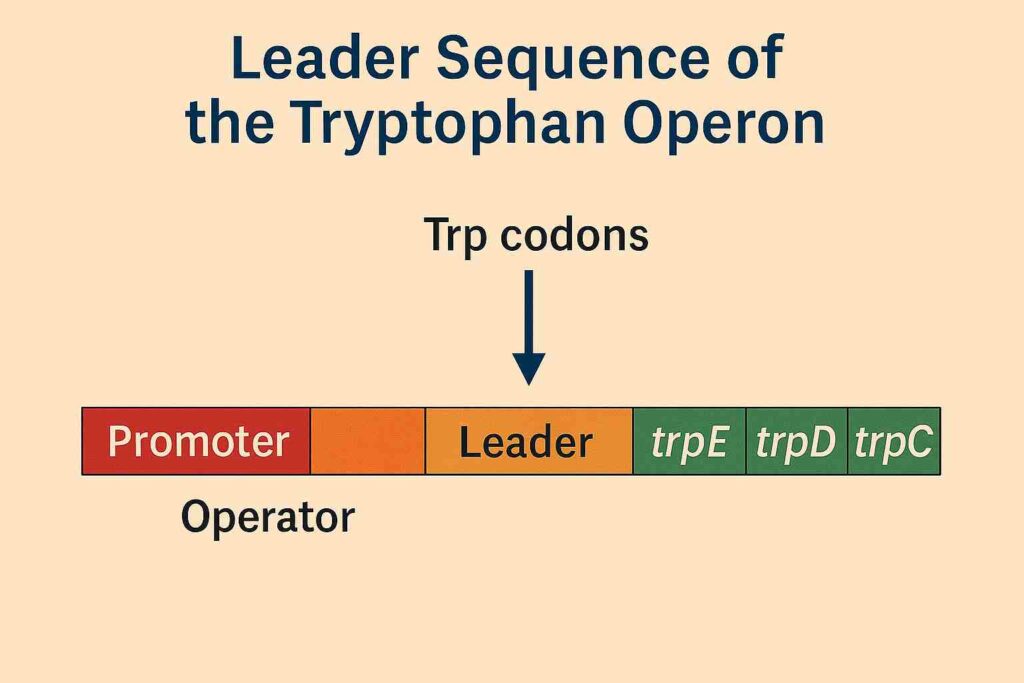
Between the promoter-operator region and the first structural gene lies the leader sequence (trpL). This region plays a crucial role in an additional regulatory mechanism known as attenuation, which fine-tunes gene expression based on tryptophan availability. The leader sequence includes a short coding region with two tryptophan codons and a series of inverted repeats capable of forming alternative secondary structures in the mRNA.
Function of the Tryptophan Operon
The Tryptophan (trp) operon plays a crucial role in regulating the synthesis of the amino acid tryptophan in prokaryotic organisms such as Escherichia coli. Its primary function is to ensure that tryptophan is synthesized only when it is not available from the environment, thereby conserving energy and resources within the cell.
Regulation of Tryptophan Biosynthesis
The most fundamental function of the trp operon is to control the biosynthesis of tryptophan. The operon contains structural genes encoding enzymes necessary for the stepwise conversion of chorismic acid to tryptophan. These enzymes are:
- Anthranilate synthase (trpE, trpD)
- Phosphoribosyl anthranilate transferase (trpC)
- Indole-3-glycerol phosphate synthase (trpC)
- Tryptophan synthase β-subunit (trpB)
- Tryptophan synthase α-subunit (trpA)
When intracellular tryptophan levels are low, the operon ensures the continuous production of these enzymes to meet the cell’s metabolic needs.
Energy and Resource Conservation
Another essential function of the trp operon is to promote metabolic efficiency by turning off the biosynthetic pathway when tryptophan is sufficiently abundant. The biosynthesis of amino acids like tryptophan requires considerable energy and molecular resources. By downregulating the operon in the presence of adequate tryptophan, the cell conserves ATP, precursor metabolites, and other resources for use in other vital cellular processes.
Negative Feedback Regulation
The trp operon functions as a classic example of negative feedback regulation in prokaryotic gene expression. When tryptophan accumulates in the cytoplasm, it binds to the inactive trp repressor protein, activating it. The active repressor then binds to the operator region of the operon, physically blocking the transcription of structural genes. This prevents the unnecessary production of tryptophan-synthesizing enzymes.
This feedback mechanism ensures a stable internal concentration of tryptophan, preventing both deficiency and wasteful overproduction.
Fine-Tuning Gene Expression via Attenuation
In addition to repression, the trp operon employs an elegant attenuation mechanism for fine-tuning gene expression. The leader sequence (trpL) located between the operator and the first structural gene contains a short peptide-coding region rich in tryptophan codons and specific sequences capable of forming stem-loop structures in the mRNA.
When tryptophan is abundant, ribosomes quickly translate the leader peptide, allowing the formation of a terminator hairpin structure in the mRNA. This structure halts transcription before the structural genes are transcribed. If tryptophan is scarce, ribosomes stall at tryptophan codons, leading to the formation of an anti-terminator structure and continuation of transcription.
Thus, attenuation allows rapid, responsive adjustment of enzyme production based on subtle fluctuations in tryptophan levels.
Coordination of Polygenic Expression
The trp operon organizes multiple genes under a single promoter-operator system, enabling the coordinated expression of functionally related enzymes. All five structural genes are transcribed into a single polycistronic mRNA molecule, ensuring that enzymes required for sequential steps in tryptophan biosynthesis are produced together, in equal proportion, and under the same regulatory control.
This organization is particularly efficient in prokaryotic cells, where the simultaneous regulation of related genes helps maintain balanced metabolic pathways.
Evolutionary and Experimental Significance
Beyond its physiological roles, the trp operon serves as a vital experimental model in molecular biology research. It has been extensively used to investigate fundamental principles of gene regulation, protein-DNA interactions, transcriptional control, and metabolic feedback loops.
Its functions have also provided insights into the evolution of gene regulation strategies in bacteria and the origin of complex regulatory mechanisms in higher organisms.
Regulation of the Tryptophan Operon
Negative Feedback Control
The trp operon operates under a negative feedback control mechanism, where the end product of the metabolic pathway, tryptophan, inhibits its own synthesis by affecting gene transcription. As tryptophan levels rise, it binds to the inactive trp repressor protein, converting it into its active form. The active repressor then attaches to the operator region, blocking RNA polymerase from transcribing the structural genes.
Role of the Repressor Protein
The trp repressor protein is synthesized constitutively by the trpR gene but remains inactive unless bound by tryptophan. In the absence of tryptophan, the repressor cannot attach to the operator, and transcription proceeds uninterrupted. When tryptophan molecules are present, they serve as corepressors by binding to the repressor protein, altering its conformation and enabling it to bind to the operator, thus halting transcription.
Attenuation Mechanism
Apart from repression via the repressor protein, the trp operon features an additional regulatory mechanism known as attenuation, which fine-tunes gene expression based on tryptophan availability during the early stages of transcription. The leader sequence of the mRNA forms alternative secondary structures called stem-loop hairpins that determine whether transcription will continue or terminate prematurely.
When tryptophan is abundant, the ribosome quickly translates the leader peptide, allowing the formation of a transcription termination loop (3-4 stem-loop), causing RNA polymerase to disengage before transcribing the structural genes. When tryptophan is scarce, the ribosome stalls at the tryptophan codons in the leader peptide, leading to the formation of an antiterminator loop (2-3 stem-loop), which permits the continuation of transcription.
Biological Significance of the Tryptophan Operon
Efficient Resource Management
The trp operon exemplifies a resource-efficient regulatory system. By repressing tryptophan synthesis when sufficient levels are present, the cell avoids wasting energy and resources producing unnecessary enzymes. This adaptive response is vital for survival, especially in fluctuating environmental conditions.
Insight into Prokaryotic Gene Regulation
The trp operon serves as a model system for understanding prokaryotic gene regulation. Its dual regulatory mechanisms — repression by the repressor protein and attenuation via mRNA secondary structures illustrate the sophisticated strategies employed by even simple organisms to control gene expression with precision.
Applications in Genetic Research
Studying the trp operon has provided valuable insights into the principles of transcriptional control, operon organization, feedback inhibition, and molecular genetics. It remains a central topic in genetics, molecular biology, and biotechnology, contributing to the development of gene regulation models and genetic engineering applications.
Research Significance of the Tryptophan Operon
Research on the trp operon has significantly contributed to our understanding of gene regulation:
Discovery of Attenuation
The concept of attenuation was first described through the study of the trp operon, marking a significant milestone in molecular biology. This demonstrated how translation can influence transcription in prokaryotes.
Regulation of Amino Acid Biosynthesis
By studying the trp operon, scientists gained insights into how bacteria efficiently regulate amino acid biosynthesis in response to environmental and intracellular conditions.
Evolutionary Insights
Comparative studies of operons like the trp operon across different bacterial species provide clues about the evolutionary conservation and diversity of gene regulation mechanisms.
Difference Between Lac Operon and Tryptophan Operon
Though both the lac operon and trp operon are classic prokaryotic operons regulating gene expression in E. coli, they differ in structure, function, and regulation.
Functional Role
The lac operon controls the metabolism of lactose, a disaccharide sugar, by producing enzymes needed to break it down. In contrast, the trp operon regulates the biosynthesis of tryptophan, an essential amino acid.
Type of Regulation
The lac operon is an inducible operon, usually off but can be turned on in the presence of lactose. Lactose acts as an inducer by binding to the lac repressor, inactivating it and allowing transcription.
The trp operon is a repressible operon, typically on but can be turned off when tryptophan is abundant. Tryptophan acts as a corepressor, activating the trp repressor to bind the operator and halt transcription.
Control Mechanism
The lac operon uses induction and involves the binding of the inducer (allolactose) to the repressor, whereas the trp operon uses feedback repression, where tryptophan binds to the repressor to inhibit its own synthesis.
Presence of Attenuation
Another key difference is that the trp operon uses attenuation as an additional regulatory mechanism, a feature absent in the lac operon.
Number of Structural Genes
The lac operon comprises three structural genes:
- lacZ
- lacY
- lacA
The trp operon includes five structural genes:
- trpE
- trpD
- trpC
- trpB
- trpA
End Product
The end product of the lac operon is the metabolism of lactose into glucose and galactose, while the trp operon results in the synthesis of tryptophan.
Application of the Tryptophan Operon
The trp operon serves as a fundamental model in molecular biology and biotechnology for studying gene regulation mechanisms. Several applications include:
Genetic Engineering
Understanding the trp operon has allowed researchers to develop synthetic operons and regulatory systems for controlled gene expression in bacterial systems, useful in industrial microbiology and pharmaceutical protein production.
Antibiotic Research
Since bacteria rely on pathways like tryptophan biosynthesis, which are absent in humans, components of the trp operon pathway are targets for designing antibacterial agents that inhibit bacterial growth without affecting human cells.
Molecular Genetics Education
The trp operon is commonly used in teaching to explain how feedback inhibition and gene regulation work in prokaryotic organisms, often contrasted with other operons like the lac operon.
Conclusion
The tryptophan operon represents an elegant example of genetic regulation through a repressible operon system in prokaryotes. It controls the biosynthesis of tryptophan via negative feedback mechanisms involving both repression and attenuation. The interplay between the repressor protein, corepressor molecule, operator region, and leader sequence ensures that tryptophan synthesis occurs only when necessary. The operon not only ensures efficient resource management within bacterial cells but also provides a classic model for studying gene regulation, making it a cornerstone topic in molecular genetics.
Frequently Asked Questions (FAQ)
What is the tryptophan operon?
The tryptophan operon is a repressible gene system in E. coli that controls the production of enzymes needed for tryptophan biosynthesis.
What is the difference between the lac operon and the tryptophan operon?
The lac operon is inducible, activated in the presence of lactose, while the tryptophan operon is repressible, turned off when tryptophan is abundant.
What is the Trp repressor in E. coli?
The Trp repressor is a regulatory protein in E. coli that binds to the operator of the trp operon in the presence of tryptophan, blocking gene transcription.
Related Contents
DNA Replication – The Molecular Mechanism of Genetic Inheritance
Phylum Echinodermata: General Characteristics and Classification