Worldwide recognition exists for the genus Staphylococcus spp. as the cause of bacterial illnesses in both humans and animals. Antimicrobial resistance may rise when dairy animals treated with antibiotics and insufficient management. This study aimed to characterize 95 strains of Staphylococcus spp bacteria that were isolated from both conventional and organic Minas Frescal cheese manufacturing. The strains were selected based on their antibiotic resistance (phenotype and genotype), presence of biofilm-formation and sanitizer-resistant genes, and SCCmec typing.
Table of Contents
Description (Staphylococcus spp)
The majority of strains (25.3%) had greater penicillin resistance, with oxacillin (21.1%) and clindamycin (11.6%) following closely behind. The most common genes for antibiotic resistance were blaZ (25.3%), mecA (13.7%), lsaB (6.3%), msrA (4.2%), ant4 (3.2%), and tetM (2.1%); the most common genes for sanitizer resistance were qacA/B (5.3%) and qacC (6.3%); the most common genes for biofilm were bap (4.2%), icaA (29.5%), and icaD (41.1%).
The lack of synthetic antibiotic use on conventional farms during the sample collection period may have contributed to the lack of a statistically significant difference between dairy products from organic and conventional farms. Methicillin-resistant coagulase-negative staphylococci possessed nontypeable SCCmec, while methicillin-resistant Staphylococcus aureus (MRSA) had kinds I and IVc SCCmec.
These findings imply that the production of Minas Frescal cheese in the Brazilian state of São Paulo, both conventional and organic, contains antibiotic-resistant bacteria. This bolsters the notion that better quality control is required all the way from the point of milking to the finished product.
Another common source of infections in humans is Staphylococcus. Symbionts in the skin, glands, and mucous membranes, they can also cause pathogenic conditions that lead to cellulite, impetigo, boils, pneumonia, bacteremia, osteomyelitis, encephalitis, meningitis, chorioamnionitis, scalded skin syndrome, abscesses in the muscles, and infections of the urogenital and abdominal tracts. For expressing different pathogenic pathways, Staphylococcus aureus has long been regarded as the most harmful species (Fontana and Favaro, 2018); nevertheless, in recent years, CNS have generated interest as possible sources of nosocomial infections (Martínez-Meléndez et al., 2015).
Resistant to methicillin The primary human infections with increased virulence and drug resistance are Staphylococcus spp aureus (MRSA). MRSA can be further split into community-associated MRSA and healthcare-associated MRSA. MRSA can cause a variety of infections, ranging from mild to severe illnesses (Wang et al., 2010; Barcudi et al., 2020). (Wang et al., 2010; Yanagihara et al., 2012). Notably, methicillin resistance can also develop in the central nervous system (CNS) (Martínez-Meléndez et al., 2015). Accordingly, in both nosocomial and community-acquired infections, staphylococci of any kind may be cause for worry (Fontana and Favaro, 2018).
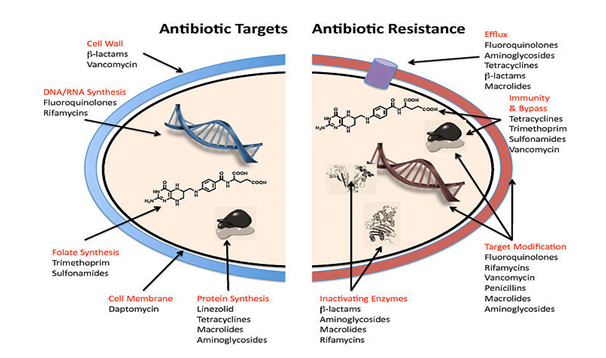
Here are some of the key antibiotic resistance genes found in Staphylococcus spp species:
mecA:
- Methicillin and other beta-lactam antibiotics are resistant.
- Mechanism: Produces a modified penicillin-binding protein (PBP2a) that has a low beta-lactam affinity.
- Frequent in: Staphylococcus aureus resistant to methicillin (MRSA).
vanA and vanB:
- Resistance: Vancomycin.
- Mechanism: Alters the peptidoglycan synthesis pathway, leading to reduced binding of vancomycin.
- Common in: Vancomycin-resistant Staphylococcus aureus (VRSA).
ermA, ermB, and ermC:
- Resistance: Macrolides, lincosamides, and streptogramin B (MLSB).
- Mechanism: Methylation of 23S rRNA, preventing binding of these antibiotics.
- Common in: Both S. aureus and coagulase-negative staphylococci.
tetK and tetM:
- Resistance: Tetracyclines.
- Mechanism: Efflux pump (tetK) and ribosomal protection protein (tetM).
- Common in: Various Staphylococcus species.
aac(6′)-Ie-aph(2”)-Ia:
- Resistance: Aminoglycosides (e.g., gentamicin).
- Mechanism: Enzyme-mediated modification (acetylation and phosphorylation) of the antibiotic.
- Common in: Both S. aureus and coagulase-negative staphylococci.
blaZ:
- Resistance: Penicillin.
- Mechanism: Beta-lactamase production, which hydrolyzes the beta-lactam ring of penicillin.
- Common in: Penicillin-resistant Staphylococcus aureus (PRSA).
qacA and qacB:
- Resistance: Quaternary ammonium compounds (disinfectants) and some antibiotics.
- Mechanism: Efflux pumps that expel toxic substances from the cell.
- Common in: Various Staphylococcus species, often associated with hospital environments.
Bacteria can easily spread these resistance genes by finding them on plasmids, transposons, or chromosomal DNA. One important contributing element to the spread of antibiotic resistance among staphylococci is this horizontal gene transfer.
Result
Public health is facing an increasing difficulty as the prevalence of antibiotic-resistant infections has risen and the approval of new medications has reduced. Nonetheless, agricultural animals are important reservoirs for resistant pathogens and can spread them through food items; as a result, the use of antibiotics in these animals has been suggested as a means of selecting resistant microbes (Nordstrom et al., 2013; FDA, 2020).
The rise of resistant bacteria, which carry resistance genes and can infect food, animals, and humans as well as transfer their genes to other bacteria, is a result of the usage of antimicrobials in food production. resistant to methicillin Among microorganisms that are resistant to antibiotics, Staphylococcus spp aureus stands out.
It has been detected in food-producing animals and those who come into touch with them, and it is currently prevalent in many hospitals across the world. (Kluytmans, 2010).
Resistant to methicillin The mec gene (mecA, mecB, and mecC) confers resistance to β-lactam antibiotics in Staphylococcus spp aureus by encoding a novel penicillin-binding protein (PBP2a; Liu et al., 2016). Methicillin resistance is made possible by the penicillin-binding protein (PBP2a or PBP2), which is encoded by the mecA gene (Wang et al., 2015). According to Wan and Chou (2014), these binding proteins reduce the potency of β-lactam antibiotics against bacteria.
The SCCmec element functions as a carrier for the exchange of genetic information between Staphylococcus spp strains and carries mecA, mecB, and mecC as well as the genes that regulate its expression, mecR1 (encoding the MecR1 protein-transducing signal) and MecI (encoding the MecI-repressor protein) (Liu et al., 2016). Moreover, SCCmec contains site-specific recombinases known as cassette chromosomal recombinases (ccr), which are in charge of each element’s mobility. Three different ccr gene types—ccrA, ccrB, and ccrC—have already been found in S. aureus (Doulgeraki et al., 2017). There are currently 11 recognized forms of SCCmec (IWG-SCC, 2014).
This study’s goals were to perform SCCmec typing of the MRSA samples discovered to observe the microbiological differences between the two cheese production methods, and to characterize Staphylococcus spp. isolates in organic and conventional Minas Frescal cheese production in the state of São Paulo, Brazil regarding antibiotic resistance, sanitizer-resistance genes, and biofilm production.
Frequently Asked Question(FAQ)
What are the antibiotic resistance genes in Staphylococcus?
Another gene linked to staphylococci’s spp resistance to penicillin is blaZ, encoding the enzyme β-lactamase 2. In order to stop the spread of infections, it is critical to identify antibiotic resistance genes quickly and accurately while treating staphylococcal infections.
What is an antibiotic resistant strain of staph?
Skin infections caused by Staphylococcus spp aureus, sometimes known as “staph,” are becoming more common and are resistant to many antibiotics (drugs that kill bacteria). Methicillin-resistant Staphylococcus sppaureus, or “MRSA,” is the term used to describe these resistant staph strains.
What are the names of the antibiotic resistance genes?
Selectable marker genes that are resistant to antibiotics help in the process of identifying host cells that harbor the vector (transformants) and removing non-transformants. Tetracycline and ampicillin are two examples of antibiotic resistance genes that can be employed as E. coli selectable markers.
Related Article